Introduction
Different types of restriction enzymes are present in the molecular biology world. Restriction enzymes come under the class of nucleases. They are termed molecular scissors for manipulating DNA.
Hence, these restriction endonucleases are responsible for cutting DNA at specific target sites known as recognition sites.
Classification of restriction enzymes based on recognition sites
Restriction enzymes are broadly classified into Type I, II, III, and Type IV enzymes.
Type I Restriction Enzymes
Description
Type I restriction endonucleases are complex enzymes and have a recognition sequence of about 15 base pairs. They cleave the DNA about 1000bp away from the 5′ – end of the sequence.
Recognition site
The type 1 target site is a specific recognition site in a DNA molecule that is easily recognizable by proteins. They are important for gene regulation and gene expression in a cell.
Type 1 restriction enzymes contain 8-12 base pairs that are found in humans. It contains palindromic sequences and hence makes them easier for protein binding.
Cleavage and mechanism
The cleavage mechanism involves two steps:
- Binding– The enzyme binds to its target sequence on one strand of double-stranded DNA.
- Cutting– Then it cuts both strands at their target sites. Once cut, new genetic material is inserted by cleaving these sites.
Both these processes are widely used in molecular biology research and biotechnology applications like gene therapy and gene cloning. It thus plays a key role in learning the evolution of an organism over time
Applications
- In DNA cloning and sequencing.
- Genome editing– Restriction enzymes help in the manipulation of genomes by removing and replacing specific sites of DNA. This process is called gene targeting.
- Diagnostics research– Techniques like PCR are used to detect genetic diseases.
Type II Restriction Enzymes
Description
Type II restriction endonucleases are stable and induce cleavage inside or outside immediately in their recognition site.
Type II enzymes require Mg²+ ions for cleavage. The first type II restriction enzyme HindII was extensively used in gene mapping and gene cloning at specific sites. HindIII is an example.
Recognition Site
The recognition sequences for type II endonucleases form palindromes with rotational symmetry. In a palindrome, the base sequence in the second half of a DNA strand is the mirror image of the first sequence.
The complementary DNA strand of a double helix is the second half of its complementary strand. Most of the type II restriction enzymes have recognition sites of 4, 5, or 6 bp.
Cleavage Patterns and Mechanisms
Most of the type II restriction endonucleases cleave the DNA molecule within their specific recognition sequences. E.g., NlaIII, Sau3A, etc.
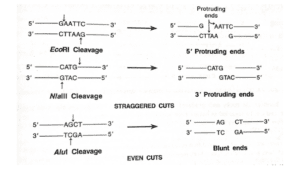
They easily pair with sticky ends which is an important property for the construction of recombinant DNAs.
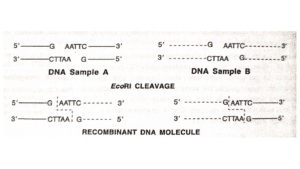
Applications
- DNA sequencing and analysis.
- They cut DNA into smaller fragments that are used for analysis for various purposes.
- In Genetic engineering and gene therapy research.
- In Diagnostic testing and forensics investigation for quick analysis.
Type III Restriction Enzymes
Description
Type III restriction endonucleases are an intermediate between type I and type II enzymes. They are responsible for cleaving DNA in the immediate vicinity of the recognition site. The recognition site for this enzyme is 4 – 8 base pairs long.
Recognition Sequence
They recognize asymmetric target sites and cleave the DNA on one side of the recognition sequence upto twenty base pairs. EcoP1, EcoP15 and HinfIII are examples of type III restriction enzymes.
Type III restriction enzymes have two units:
- Site-specific endonuclease (REase)
- coordinating methyltransferase (MTase).
The site-specific restriction enzyme helps in recognizing the target by binding with the help of hydrogen bonds between them.
After binding, it cleaves both the DNA strands either at or near their recognition sites. They generally exhibit palindromic properties.
Hence, it allows easy binding as anyway they face parallel to each other.
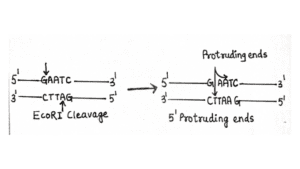
Cleavage and Mechanism
They cleave DNA at specific target sites in the DNA. The latter fragments are then used for many purposes that include cloning, amplification, and gene expression analysis.
The process of cleavage is based on certain criteria
- Concentration of enzymes
- Temperature
- Salt concentration
- pH level
- Substrate sequence length.
These enzymes require high concentrations to work because of their lower affinity towards the target sequences. Their high sensitivity to environmental changes affects their activity levels.
Applications
- Type 3 restriction enzymes are used in DNA sequencing.
- They have proved to be more efficient in recognizing specific site sequences in DNA molecules.
- They provide insights and effective interaction with different substrates that help in manipulating the gene expression in cells.
Type IV restriction enzymes
Description
Type IV restriction enzymes are used to cut DNA at specific sites. They help in recognizing and binding to short sequences in the DNA.
Recognition site
Recognition sites of Type IV restriction enzymes include:
- EcoRI – 5’-GAATTC-3’
- BamHI – 5’-GGATCC-3’
- HindIII – 5′-AAGCTT-3′
- XhoI – 5′-CTCGAG-3′
Cleavage and Mechanism
These enzymes belong to a group of DNA-cleaving proteins that cleave specific sequences in ds- DNA. They are hence widely used in molecular applications, like gene cloning and DNA sequencing.
Cleavage is dependent on various factors, including:
- Recognition of the targeted site
- Presence of metal ions
- Balanced pH levels
- Regulation of temperature
- Environmental conditions.
Type IV restriction endonucleases are used for research purposes by combining the different sequence sites together.
Applications
- Analysis of DNA sequence– The sequence is analyzed to verify the genomic information.
- Genetic engineering– Insertion of a new DNA sequence in order to achieve modification and correction of a gene defect.
- Gene Cloning – Creating copies of a particular genome so as to correct hereditary diseases.
- Diagnostics research – To detect genetic disorders in an individual and diseases like HIV etc.
Applications of restriction enzymes
1. DNA cloning
Basically, a recombinant DNA molecule is a vector into which a desired DNA fragment has been inserted to enable its cloning in an appropriate host.
2. In PCR
PCR is used to amplify a specific gene present in different individuals of a species including somatic cells such as human sperm.
These data can be sequenced to obtain information on the mutational changes in the genes of individuals. They are used for disease diagnosis, population genetic studies, etc.
PCR is applied for prenatal diagnosis of genetic diseases. Example: sickle cell anemia
The technique of PCR is also used to determine the sex of the embryos. Thus the sex of in vitro fertilized cattle embryos is by Y-chromosome-specific primers before implantation.
RT-PCR provides information on the activity of tumor cells and viruses.
PCR also assists in the mapping of human genome sequences.
3. In DNA fragment analysis
The identification and isolation of the desired DNA fragment is a critical step in recombinant DNA technology.
A mix of DNA fragments is used to construct a genomic library. cDNA obtained from mRNA are cloned to give rise to the cDNA library.
When amino acid sequences of a gene are known, they are either synthesized chemically or by using a polymerase chain reaction.
Restriction enzymes and emerging technology
Restriction enzymes in emerging technologies have become very popular. This progressed because of the following factors:
- They are widely used for cloning and manipulation of DNA that are used to discover therapies for incurable diseases.
- These enzymes are used in DNA sequencing programs, allowing researchers to understand genetic mutations in a better manner.
- The introduction of CRISPR-Cas9 technologies simplified the idea of gene manipulation using restriction enzymes.
Creating artificial DNA
- Artificial DNA is basically a creation through synthetic biology that works by manipulating genetic material. Recently, it became popular due to its applications like gene therapy (correction of a gene defect ), and agricultural biotechnology to get rid of weeds.
- The technique involves the creation of new DNA strands from existing DNA and modifying them so as to achieve the desired artificial DNA.
- Many advantages are associated with artificial DNA creation by manipulation of genes like rDNA technology. This process thus enables high accuracy and precision with faster result generation.
Programmable DNA cleavage
Programmable DNA cleavage is a methodology that allows genetic researchers to control the cleaving in DNA. It includes gene manipulation and regulation of genes.
The basic idea behind DNA cleavage is high cleaving precision in an efficient manner compared to other methods.
The necessity of programmable DNA cleavage has increased in recent years because of advancements in molecular biology techniques. Moreover, the development of new enzymatic tools made it easier for the researchers to design customized enzymes with high accuracy and increased specificity.
Hence, programmable DNA cleavage is an important tool in manipulating genetic material with accuracy.
Q&A
1. How many types of restriction enzymes are there?
There are basically four types of restriction enzymes.
2. What are the different types of restriction enzymes?
Restriction enzymes are broadly classified into Type I, II, III, and Type IV enzymes.
3. What are the three types of restriction enzymes?
The three types of restriction enzymes include Type I, Type II, and Type III.
Type I restriction endonucleases are complex enzymes and have a recognition sequence of about 15 base pairs. They cleave the DNA about 1000bp away from the 5′ – end of the sequence.
Type II enzymes require Mg²+ ions for cleavage. The first type II restriction enzyme HindII was extensively used in gene mapping and gene cloning at specific sites. HindIII is an example.
Type III restriction endonucleases are an intermediate between type I and type II enzymes. They are responsible for cleaving DNA in the immediate vicinity of the recognition site. The recognition site for this enzyme is 4–8 base pairs long.
4. What are the two types of restriction enzymes?
Ans: Two types of restriction enzymes include: Type I and Type II.
Type I restriction endonucleases are complex enzymes and have a recognition sequence of about 15 base pairs. They cleave the DNA about 1000bp away from the 5′ – end of the sequence.
Type II enzymes require Mg²+ ions for cleavage. The first type II restriction enzyme HindII was extensively used in gene mapping and gene cloning at specific sites.
5. What are the types of restriction enzymes?
Ans: There are four different types of restriction enzymes that cleave DNA molecules. They are classified as Type I, II, III, and Type IV enzymes.
Summary
- Different types of restriction enzymes that cleave the DNA molecule like Type I, II, III, and Type IV.
- Restriction enzymes come under the class of nucleases.
- They are termed molecular scissors for manipulating DNA.
- Hence, these restriction endonucleases are responsible for cutting DNA at specific target sites known as recognition sites.
- Type I restriction endonucleases are complex enzymes and have a recognition sequence of about 15 base pairs.
- Type II enzymes require Mg²+ ions for cleavage. The first type II restriction enzyme was HindII which was extensively used in gene mapping
- Type III restriction endonucleases are an intermediate between type I and type II enzymes.
- They are responsible for cleaving DNA in the immediate vicinity of the recognition site. The recognition site for this enzyme is 4–8 base pairs long.
- Type IV restriction enzymes are used to cut DNA at specific sites. They help in recognizing and binding to short sequences in the DNA.
- Restriction enzymes in emerging technologies have become very popular.
References
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC331348/
- https://febs.onlinelibrary.wiley.com/doi/abs/10.1111/j.1432-1033.1997.t01-6-00001.x
- https://link.springer.com/article/10.1007/s12038-019-9856-8
- https://www.cell.com/biophysj/pdf/S0006-3495(21)01354-0.pdf
- https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0006441
