Introduction
HindIII restriction enzyme is a molecular scissors that helps in cleaving DNA molecules. The term genetic engineering is a technique for modifying organisms through heredity and reproduction. It includes a wide range of biomedical techniques such as artificial insemination, in vitro fertilization, and gene manipulation.
Restriction endonucleases come under nucleases. They are molecular scissors for manipulating DNA. Hence are responsible for cutting DNA at specific sites known as recognition sites.
Hind III is an example of a type II restriction enzyme.
HindIII is a type II restriction endonuclease. Stable and helps in cleavage in the presence of Mg²+ ions. They are responsible for restriction mapping and gene cloning that cleaves at specific target sites. Type II restriction enzymes such as HindIII are also used in gene cloning.
The world of restriction enzymes
Restriction enzymes were postulated by Werner Arber in the 1960s and were responsible for cleaving DNA molecules at specific recognition sites. The recognition sites are different and specific for different restriction enzymes. The recognition sequences for HindIII is A/AGCTT
They are an essential tool for DNA cloning and sequencing. They serve as the tools for cutting DNA molecules at predetermined sites, that is the basic requirement for gene cloning of recombinant DNA technology.
Role of Restriction Enzymes
- Restriction enzymes act as a ‘protection’ system for bacteria.
- They hydrolyze exogenous DNA molecules that are not methylated by the host modification enzyme.
- These enzymes belong to a group called nucleases and they cut at an internal position in a DNA strand. Hence, they are known as endonucleases.
- Restriction enzymes are derived from the bacterium from which their strains are isolated.
- Restriction enzymes can generate ‘sticky’ ends that can be used to join DNA from two different sources together for the generation of rDNA molecules.
Diversity
Restriction enzymes are broadly classified into Type I, II, III and Type IIs enzymes.
Type I restriction endonucleases are complex enzymes and have a recognition sequence of about 15 base pairs. They cleave the DNA about 1000bp away from the 5′ – end of the sequence.
Type II restriction endonucleases are stable and induce cleavage inside or outside immediately in their recognition site.
Type II enzymes require Mg²+ ions for cleavage. The first type II restriction enzyme HindII was extensively used in gene mapping and gene cloning at specific sites. HindIII is an example.
Type III restriction endonucleases are an intermediate between type I and type II enzymes. They are responsible for cleaving DNA in the immediate vicinity of the recognition site.
They recognize asymmetric target sites and cleave the DNA on one side of the recognition sequence up to twenty base pairs. EcoP1, EcoP15, and HinfIII are examples of type III restriction enzymes.
HindIII restriction enzymes
History
HindIII is a Restriction Endonuclease that is obtained by purifying from a bacteria Escherichia coli strain that carries the HindIII gene from Haemophilus influenza Rd at an optimum temperature of 37°C.
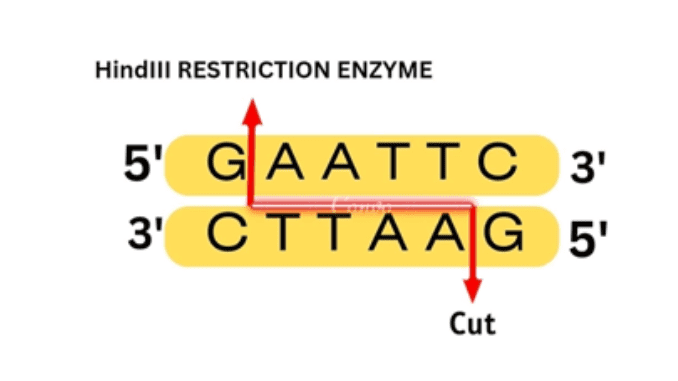
It is not affected by Dam methylation and follows a palindromic sequence 5′-AAGCTT- 3′. HindIII restriction enzyme required Mg²+ for restriction.
They have the ability to recognize specific sites within the DNA but do not cut these sites. The location of endonuclease and methylase are on a single protein molecule.
Source of organisms
Haemophilus influenza Rd
The nomenclature of restriction enzymes follows a general pattern. The first letter of the name of the genus in which a given enzyme is discovered is capital. Hin is derived from Haemophilus influenzae.
When a particular bacterium produces more than one strain, they are written as roman numerals. The different enzymes produced by the H. influenzae strain Rd are named HindII, and HindIII respectively.
Function and mechanism of action
How does this enzyme function as molecular scissors?
Hind III was considered as the third of four enzymes that were extracted from Haemophilus influenzae. They act as molecular scissors to cut at specific sites known as recognition sites.
Recognition sequence
The recognition sequences for HindIII restriction enzymes form palindromes with rotational symmetry. The base sequence in the first half of one strand of a DNA double helix is the mirror image of its second half mirror image of the second half its complementary strand.
Thus in such palindromes, the base sequence in both the strands of a DNA duplex reads the same when read from the same end either 5′ or 3′ of both the strands.
Most of the type II restriction enzymes have recognition sites of 4, 5, or 6 base pairs. Few restriction enzymes have uncertainties in their recognition sites, like Hinfl, and EcoRII, which may contain recognition sites up to 4 base pairs.
Cleavage Patterns
Most type II restriction endonucleases cleave the DNA molecules within their specific recognition sequences, but some produce cuts immediately outside the target sequence, such as HindIII
Most enzymes create cuts where strands of DNA are cleaved at different positions. Due to the symmetrical nature of the target sites, the two protruding ends generated by such a cleavage have complementary base sequences. These ends are known as Sticky ends.
Enzyme substrate interaction of these enzymes
Recognition of genetic sequence
The enzyme-substrate interaction of HindII is a complex process involving the binding of an enzyme to a specific substrate. It generally has two base pairs that are divided into nucleotide sequences.
This helps in cleaving the DNA strands at particular locations. HindIII restriction enzymes help in catalyzing the hydrolysis between recognition sequences i.e., (5′-AAGCTT-3′).
Each strand leads to the formation of blunt ends and their products are released for further ligation into the new sequences.
Importance of palindromic sequence
- Palindrome sequences are important in type II restriction enzymes because they help in recognizing the enzyme to bind to a specific DNA substrate.
- It contains two identical strands that have the same nucleotide base pairs, allowing them to be recognized by the enzyme.
- The recognition sequence for a type II restriction endonuclease has 4-6 base pairs long.
- It should possess at least one palindrome sequence in order for the enzyme to recognize.
- Once it is attached, the enzyme can cut or modify the targeted strand at the recognition site.
Applications in genetic engineering
Genetic Engineering in Medicine
Recombinant DNA technology helped scientists to produce large amounts of hormones, immuno-proteins, and proteins involved in blood clotting and blood-cell production.
Recombinant technology helps produce therapeutic drugs that can fight against certain viruses and cancers.
Gene Therapy
Gene therapy is a procedure to correct a gene defect. Here, the researchers use vectors as a medium to insert a functional gene to resolve a defective gene.
Genes are inserted into the body of the patient to correct certain hereditary diseases. The first clinical gene therapy was given in 1990 to a four-year-old girl with adenosine deaminase deficiency, which is curable by enzyme replacement therapy.
Genetically Engineered Humulin
Human insulin, also called humulin consists of two short polypeptide chains- chains A and B, linked by a disulfide bond,
Insulin is secreted as a prohormone that has to be processed before it becomes a functional hormone.
An American company prepared two DNA sequences coding chains A and B of human insulin and introduced them into the plasmids of E.coli to produce insulin.
Q&A
1. What is the HindIII restriction enzyme?
HindIII is a type II restriction endonuclease. Highly stable and helps cleavage. Require Mg²+ ions for cleavage. They are responsible for restriction mapping and gene cloning.
2. Where does the HindIII restriction enzyme cut?
The HindIII restriction enzyme cuts at 5′-AAGCTT- 3′
3. How do EcoRI and HindIII differ?
EcoRI is isolated from E. Coli bacterial species whereas HindIII is isolated from Haemophilus influenza species. They both belong to type II restriction enzymes.
4. Why is Hind II the first restriction enzyme?
HindII was isolated first and hence known as the first restriction endonuclease. On the other hand, HindI was the first restriction of exonuclease. Roman numbers depict the order from which the enzymes were isolated.
Summary
- HindIII is a type II restriction endonuclease.
- Are highly stable and help in inducing cleavage. Require Mg2+ ions for cleavage.
- Responsible for restriction mapping and gene cloning that cleaves at specific target sites.
- Type II restriction enzymes such as HindIII are also used in gene cloning.
- HindIII is isolated from Haemophilus influenza species. It belongs to type II restriction enzymes.
- The enzyme-substrate interaction of HindII is a complex process involving the binding of an enzyme to a specific substrate.
- HindIII restriction enzymes help in catalyzing the hydrolysis between recognition sequences i.e., (5′-AAGCTT-3′).
- It contains two identical strands that have the same nucleotide base pairs, allowing them to be recognized by the enzyme
- Each strand leads to the formation of blunt ends and their products are released for further ligation into the new sequences.
- When a particular bacterium produces more than one strain, they are written as roman numerals.
- The different enzymes produced by the H. influenza strain Rd are named HindII, and HindIII respectively.
- Type II restriction endonucleases cleave the DNA molecules within their specific recognition sequences, but some produce cuts immediately outside the target sequence, such as HindIII
- Once attached, the enzyme can cut or modify the targeted strand at the recognition site.
References
- https://www.sciencedirect.com/science/article/pii/0022175991900062
- https://scripts.iucr.org/cgi-bin/paper?mh5029
- https://www.sciencedirect.com/science/article/pii/002228367590232
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC310145/
- https://link.springer.com/article/10.1023/A:1024494704863
