Know in one minute about prokaryotic DNA
|
Introduction
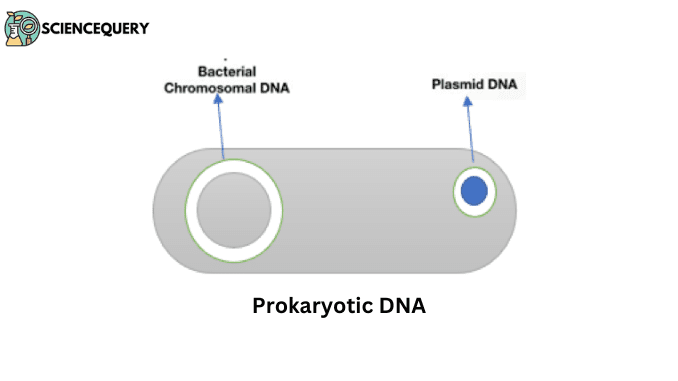
Prokaryotic DNA is a cytoplasmic circular DNA and is found in fewer amounts. This DNA is naked that is having no histones. It exists as a single chromosome, and apart from that additional plasmid DNA is also present. The compact mass of DNA is called a nucleoid.
Prokaryotic DNA replication and its steps

DNA replication is a process of forming copies of DNA. It is a semi-conservative and bidirectional process.
- Initiation
- Elongation
- Termination
Initiation
- Replication starts at a specific initiation point called the OriC point or replicon.
- In prokaryotes like E.coli the Ori C is a 245 base pair long sequence.
- DNA replication starts with the unwinding of DNA strands by the enzyme called DNA helicase. DNA helicase separates the two strands of DNA at the origin of replication and forms two Y-shaped replication forks.
- Helix destabilizing protein is attached to the single-stranded region to prevent the strands from rejoining.
- Topoisomerase reduces the tension between the strands and hence, prevents supercoiling.
- RNA polymerase forms a primer for the synthesis of the leading strand while primase is in the form of primasome synthesis for the lagging strand.
- DNA polymerase III will form the replisome. Replisomes consist of DNA polymerase and other enzymes.
Elongation
- DNA polymerase III adds nucleotides to the 5’ to 3’ direction to synthesize new strands.
- To add nucleotides free 3’-OH group is required to which DNA polymerase adds the nucleotide by forming a phosphodiester bond between the 3’-OH group end and the 5’ phosphate of the next nucleotide.
- The new strand which is complementary to the 3’ to 5’ parental strand is synthesized continuously toward the replication fork known as the leading strand.
- On the leading strand, the primer is loaded only once. While the other strand, complementary to the 5’ to 3’ parental DNA strand, is extended away from the replication fork, in small fragments called Okazaki fragments. This strand is lagging.
- On the lagging strand, primer is multiple times to form Okazaki fragments.
- Sliding clamp protein is required to hold the DNA polymerase in its place to add nucleotides continuously.
- As the synthesis of new strands proceeds, the RNA primers are replaced by DNA polymerase I, which cut the RNA and fills the gaps with DNA nucleotides.
Termination
In E. coli away from Ori C there is one large termination sequence called ‘ter’ elements that allow the two replication forks to pass through in one direction. ‘Tus’ protein binds to ter elements and stops the unwinding of DNA.
Difference between prokaryotic DNA and eukaryotic DNA
Prokaryotic DNA |
Eukaryotic DNA |
| It is located in the cytoplasm | It is located in Nucleus, mitochondria, and plastids. |
| It is circular | Linear in the nucleus and circular in mitochondria and chloroplast |
| Not associated with histone protein | Associated with histone protein |
| Forms tangled mass on denaturation | Two chains of polynucleotides appeared on denaturation |
| Codes for less number of proteins | Codes for a large number of proteins |
| Extrachromosomal DNA may be present in the form of a plasmid | Plasmid absent |
| Non-coding introns absent with little non-functional DNA | Non-coding introns present with a major part of DNA non-functional |
Difference between prokaryotic DNA replication and eukaryotic DNA replication
Prokaryotic DNA Replication |
Eukaryotic DNA Replication |
| The process of DNA replication in prokaryotes is continuous | DNA replicates in the S-phase of the cell cycle |
| Replication takes place in the cytoplasm | Replication takes place in the nucleus |
| Only one origin of replication is present | Multiple origins of replication |
| Less amount of DNA is present | The DNA is 50 times more than the prokaryotic DNA |
| DNA polymerase I and III are involved in the replication | DNA α, δ and ε are involved in replication |
| Okazaki fragments are large in size | Okazaki fragments are small in size |
| DNA gyrase is required | DNA gyrase is not required |
Q&A
1. Where is DNA located in a prokaryotic cell?
Ans. In prokaryotic cells, DNA is located in the cytoplasm.
2. What is the major difference between prokaryotic DNA replication and eukaryotic DNA replication?
Ans. The difference between prokaryotic DNA replication and eukaryotic DNA replication is given below:
| Prokaryotic DNA Replication | Eukaryotic DNA Replication |
| DNA replication process in prokaryotes is continuous | S-phase of the cell cycle is having DNA replication. |
| Replication takes place in the cytoplasm | The nucleus is the main region for replication |
| A single origin of replication is present | There are multiple origins of replication |
| Less amount of DNA is present | The DNA is 50 times more than the prokaryotic DNA |
| DNA polymerase I and III are involved in the replication | DNA α, δ and ε are involved in replication |
| Okazaki fragments are large in size | Okazaki fragments are small in size |
| DNA gyrase is required | DNA gyrase is not required |
3. Where is DNA found in prokaryotic cells?
Ans. The prokaryotic DNA found in the cytoplasm, is usually circular and present in less amount
4. Where is DNA housed in a prokaryotic cell? Where is it housed in a eukaryotic cell?
Ans. In prokaryotic cells, DNA is housed in the cytoplasm, whereas in eukaryotic cells DNA is in the nucleus.
Written By: Neetu Ladiya
Reference
Zoology – B.Sc Second year by V.K. Tiwari and V.K. Singh (Shiva Lal Agrawal and company)
