Histone acetylation: Know in one minute
a. Histone acetyltransferase (HATs) b. Histone deacetylases (HDACs).
|
Introduction
Histone acetylation is a cellular process that helps in providing binding sites for proteins thus initiating transcriptional activation.
It was first detected in the nuclei of the cell by Vincent Allfrey and his group in 1964.
Thus in simple words, we can say that a histone is present in the nuclei of the cell. And it helps in adding and deleting the acetyl group from the lysine amino acid which is present in the tail of this organelle.
Main enzymes are
- Histone acetyltransferase (HATs) enzymes
- Histone deacetylases (HDACs).
What are histones?
- Histones are a special group of proteins.
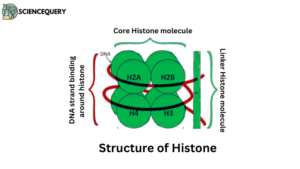
- They are highly alkaline basic proteins and positively charged, linked with negatively charged phosphate groups of DNA.
- Found in the nuclei of eukaryotic cells.
- Are responsible for DNA folding and chromatin formation.
Classes of histone
There are two main classes of histones
- Core histones
- Linker histones.
Core histones
The core histone is having four parts namely H2A, H2B which is rich in lysine, and H3 and H4 rich in arginine.
Linker histones
- Linker histone includes H1 and H5.
- H1 binds the nucleosomes at the starting and ending site of the DNA, thus locking the DNA into place and helping in the formation of a higher-order structure.
- H5 histones are used in the packaging of specific regions of DNA.
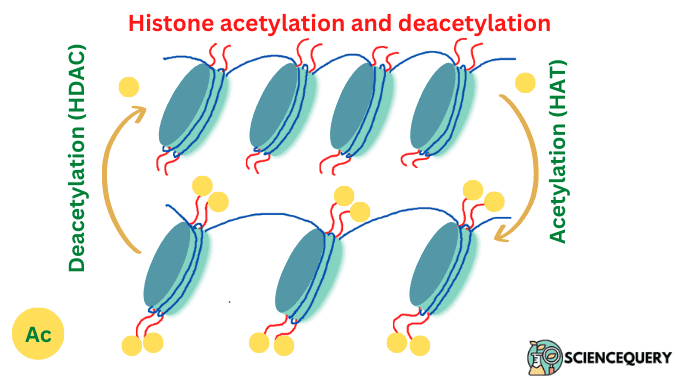
What is histone acetylation?
- Histone acetylation is a process that involves the covalent addition of an acetyl group to lysine residues in the protruding histone tails.
- It is usually associated with transcriptional activation.
- It is controlled by two enzymes namely histone acetyltransferases (HATs) which are responsible for adding acetyl groups. And histone deacetylases (HDACs) which remove them.
Histone acetyltransferases (HATs)
HATs add an acetyl group to the alpha-amino group of lysine using acetyl CoA as a cofactor, which neutralizes the positive charge on lysine, weakens the histone-DNA interaction, and makes genes accessible.
HATs are classified into two classes
- Type A
- Type-B
Type-A HATs
Type-A HATs are localized in the nucleus, and function in the transcription-related histone acetylation in chromatin.
Type-B HATs
Type-B HATs found in the cytoplasm, acetylate newly synthesized histone and influenced the structure of the nucleosome.
Histone deacetylases (HDACs)
Histone deacetylases (HDACs) are post-transcriptional modulators that remove acetyl groups from lysine residues of both histone and non-histone proteins.
Involvement in regulating fundamental cellular functions such as proliferation, cell cycle, regeneration, apoptosis, and differentiation makes them an important target of study for disorders. Deregulation of HDACs is usually associated with cancer and tumorigenesis.
Histone acetylation mechanism
- The mechanism for acetylation takes place on the amino group of lysine amino acid residue.
- These residues are located on the tails of histones that make up the nucleosomes of packaged dsDNA.
- HAT molecules facilitate the transfer of an acetyl group from a molecule of acetyl-coenzyme A to the NH₃⁺ group of lysine.
- Acetylation removes the positive charge of the histone tail, reducing affinity for the negatively charged phosphate groups of DNA.
- It also reduces the affinity of the tail for adjacent nucleosomes, thus affecting the ability of the nucleosome array to form a more repressive higher-order chromatin structure.
What is histone deacetylation?
Deacetylation is the reverse reaction where an acetyl group is removed from a molecule.
Histone deacetylases (HDACs) catalyze the removal of an acetyl group from lysine residue on histone and non-histone proteins. As a result, lysine residues retain their positive charge and histone proteins are tightly bound to DNA leading to the inhibition of gene transcription.
Functions of Histone acetylation
1. Acetylation removes the positive charge of the histone tail, reducing affinity for the negatively charged phosphate groups of DNA.
2. It also reduces the affinity of the tail for adjacent nucleosomes, thus affecting the ability of the nucleosome array to form a more repressive higher-order chromatin structure.
3. Acetylation is involved in nucleosome assembly and the interaction of histone with other regulatory proteins, creating a transcription permissive environment.
Difference between histone acetylation vs methylation
Acetylation |
Methylation |
| It is the introduction of an Acetyl functional group to the lysine amino acid of the histone tail. | It is the introduction of a methyl functional group to lysine or arginine of the histone tail. |
| Catalysis is done by using Enzymes with histone acetyltransferases (HATs) and histone deacetylases (HDACs). | These reactions are catalyzed by enzymes
with histone methyltransferase. |
| Acetylation removes the positive charge of the histone tail, reducing affinity for the negatively charged phosphate groups of DNA. | Methylation does not neutralize charge but recruits silencing or regulatory proteins that bind methylated histones. |
Q&A
1. What are histones?
Histones are a special group of proteins found in the nuclei of eukaryotic cells responsible for DNA folding and chromatin formation. Histone proteins are highly alkaline basic proteins and positively charged.
There are two main classes of histones:
- Core Histones
- Linker histones
Core histone includes
H2A-contain more lysine
H2B -contains more lysine
H3 – contains more arginine
H4 -contains more arginine
Two of each of these core histone proteins assemble to form one octameric nucleosome core particle.
Linker histone includes
H1-highest lysine/arginine ratio
H5- highest lysine/arginine ratio
H1 binds the nucleosomes at the starting and ending site of the DNA, thus locking the DNA into place and helping in the formation of a higher-order structure.
H5 histones help in packing specific regions of DNA.
2. Why do histones bind tightly to DNA?
Histones are responsible for DNA folding and chromatin formation. Histone proteins are highly alkaline basic proteins and positively charged, linked with negatively charged phosphate groups of DNA. DNA is negatively charged, due to the phosphate group and histones are positively charged and hence bind tightly.
3. Which of the following molecular characteristics cause histones to bind tightly to DNA?
Histones are alkaline or basic pH proteins, and their positive charge allows them to associate with negatively charged DNA with great affinity.
Written By: Neetu Ladiya
References
1. Molecular biology by Dr. V. K. Agrawal
2. Shivlal Unified Zoology BSc. 1st Year for MP Universities.
